Overview
Welcome to OGRP
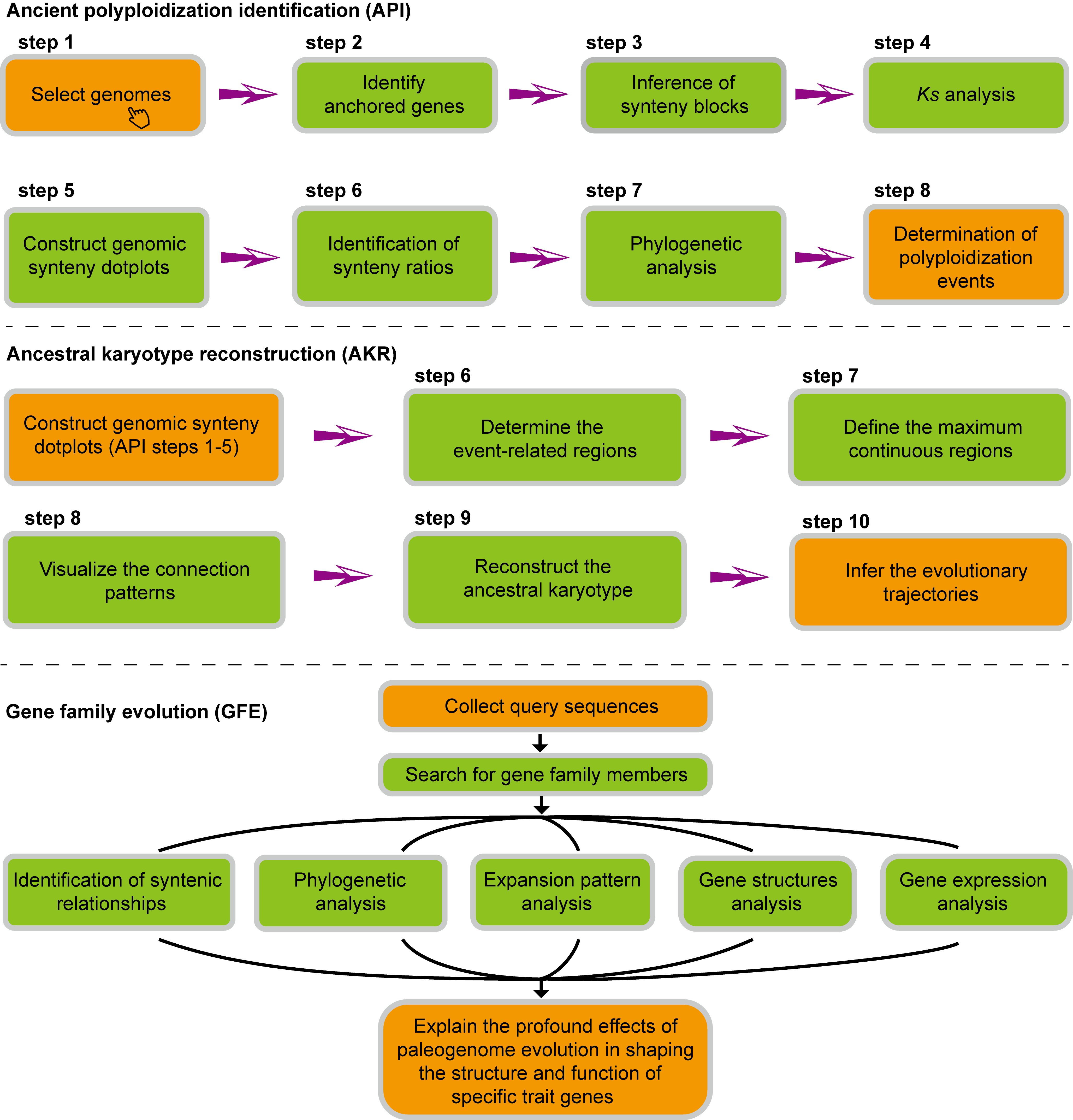
OGRP (Oleaceae Genome Research Platform) is a comprehensive and user-friendly platform designed for the functional genomics study of the Oleaceae. it is easy to obtain the 70 genomes of ten species in Oleaceae and 46 eudicots, as well as 366 transcriptomes involving 18 tissues of Oleaceae plants. We built 34 window operated bioinformatics tools (WOBTs), collected 38 professional practical software, and proposed three new pipelines: ancient polyploidization identification (API), ancestral karyotype reconstruction (AKR), and gene family evolution (GFE). Significantly, we generated a series of comparative genomic resources focusing on the Oleaceae, including 108 genomic synteny dotplots, 1,952,225 collinear gene pairs, multiple genome alignments, and imprints of paleochromosome rearrangements. Moreover, researchers can efficiently search for 1,785,987 functional annotations, 22,584 orthogroups, 29,582 important trait genes from 75 gene families, 12,664 transcription factor related genes, 9,178,872 transposable elements, and all involved regulatory pathways in Oleaceae genomes.
OGRP Portals
70
366
23,647
23,968
29,582
12,664
20,861
22,584
162,692
9,178,872
1,785,987
Genomic analysis pipelines
Genome Data (70)
| Species Name | Genome Version | Genome Access | Genome Size (Mb) | Assembly Level | Provenance |
|---|---|---|---|---|---|
| Forsythia suspensa | Forsythia-HENAU2018 | GCA_013103335.1 | 737.5 | Contig | ➠ |
| Forsythia suspensa | JALRMA01 | GCA_020510225.1 | 737.6 | Chromosome | ➠ |
| Forsythia suspensa | JAHHPY01 | GCA_023638005.1 | 737.5 | Chromosome | ➠ |
| Fraxinus americana | FRAX14-0.1 | GCA_903798225.1 | 642.6 | Scaffold | ➠ |
| Fraxinus excelsior | FRAX_001 | GCA_019097785.1 | 806.5 | Chromosome | ➠ |
| Fraxinus excelsior | BATG-0.5 | GCA_900149125.1 | 867.5 | Scaffold | ➠ |
| Fraxinus pennsylvanica | version 1.5 | GCA_912172775.2 | 756.8 | Chromosome | ➠ |
| Fraxinus pennsylvanica | FRAX09-0.1 | GCA_903798245.1 | 902.5 | Scaffold | ➠ |
| Fraxinus pennsylvanica | FRAX03-0.1 | GCA_902829185.1 | 700.9 | Contig | ➠ |
| Fraxinus pennsylvanica | FRAX10-0.1 | GCA_902829155.1 | 761 | Contig | ➠ |
| Jasminum sambac | Hutoumoli | GCA_018223645.1 | 505.5 | Chromosome | ➠ |
| Jasminum sambac | Single-petal | GWHBFHK00000000 | 502.3 | Chromosome | ➠ |
| Jasminum sambac | Double-petal | GWHBFHJ00000000 | 487 | Chromosome | ➠ |
| Jasminum sambac | ... | GWHAZHY00000000 | 550 | Chromosome | ➠ |
| Jasminum sambac | JSU-FSP | GWHBISM00000000 | 500.6 | Chromosome | ➠ |
| Jasminum sambac | Hutou | GWHDUBQ00000000 | 469.5 | Chromosome | ➠ |
| Olea europaea | O_europaea_v1 | GCA_002742605.1 | 1126.4 | Chromosome | ➠ |
| Olea europaea | Farga | GCA_902713445.1 | 1331.2 | Chromosome | ➠ |
| Olea europaea | YAF_Ocus_V1 | GCA_023089605.1 | 1228.8 | Chromosome | ➠ |
| Olea europaea | OE6 | GCA_900603015.1 | 1331.2 | Scaffold | ➠ |
| Osmanthus fragrans | Liuyejingui | GCA_019395295.1 | 733.3 | Chromosome | ➠ |
| Syringa oblata | IBSOV1 | GWHBHRY00000000 | 1146.9 | Chromosome | ➠ |
| More | ... | ... | ... | ... | ... |
Citation
If you use OGRP in your scientific research, please cite us:
・OGRP: A comprehensive bioinformatics platform for efficient empowerment of Oleaceae genomics research