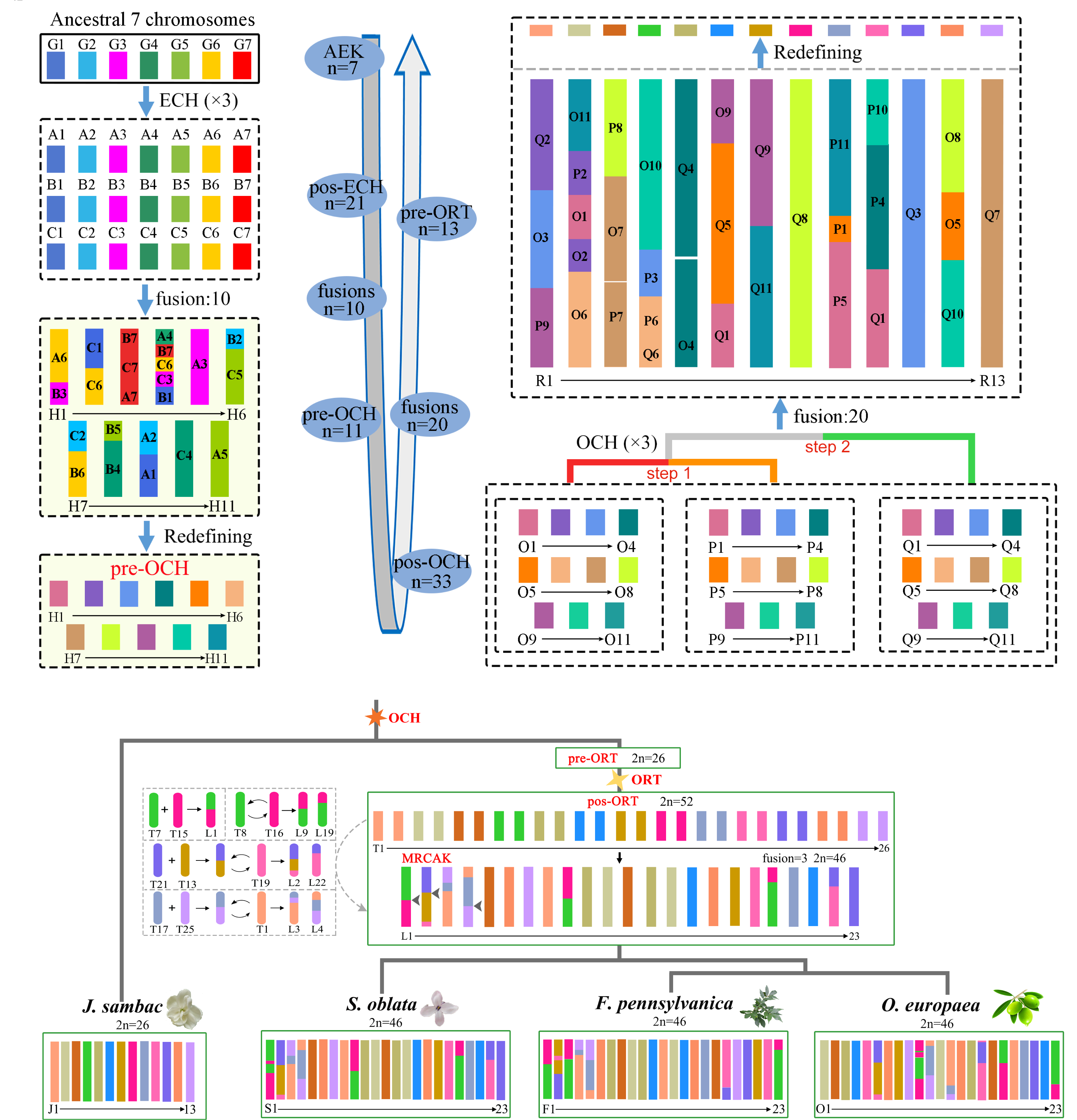
Ancestral karyotypes and evolution
We constructed the ancestral Oleaceae karyotypes of MRCAK, pre-ORT, pos-OCH, and pre-OCH and deduced the evolutionary trajectories of chromosomes in extant species. In OGRP, we created multiple animated videos that vividly present the ancestral karyotypes and evolutionary history of plants in the Oleaceae, in order to increase the value of science popularization. Of special significance, we visualized detailed genomic synteny information for all inferred ancestral karyotypes and recorded the locations of large-scale rearrangements of paleo-chromosomes preserved in the four Oleaceae genomes (O. europaea, F. pennsylvanica, S. oblata, and J.sambac) and reference genomes (C.humblotiana). Based on the inferred locations of chromosomal rearrangements (CRs), researchers can further explore the relationship between the functional evolution of important trait genes (or pathways) and CRs. As reported in previous studies, CRs enhances the plasticity of the cucurbitacin synthesis pathway, leading to the formation of key gene in the morphine synthesis pathway.
The evolutionary trajectory of Oleaceae karyotypes
Jsa 1
Jsa 2
Jsa 3
Jsa 4
Jsa 5
Jsa 6
Jsa 7
Jsa 8
Jsa 9
Jsa 10
Jsa 11
Jsa 12
Jsa 13
Sob 1
Sob 2
Sob 3
Sob 4
Sob 5
Sob 6
Sob 7
Sob 8
Sob 9
Sob 10
Sob 11
Sob 12
Sob 13
Sob 14
Sob 15
Sob 16
Sob 17
Sob 18
Sob 19
Sob 20
Sob 21
Sob 22
Sob 23
Fpe 1
Fpe 2
Fpe 3
Fpe 4
Fpe 5
Fpe 6
Fpe 7
Fpe 8
Fpe 9
Fpe 10
Fpe 11
Fpe 12
Fpe 13
Fpe 14
Fpe 15
Fpe 16
Fpe 17
Fpe 18
Fpe 19
Fpe 20
Fpe 21
Fpe 22
Fpe 23
Oeu 1
Oeu 2
Oeu 3
Oeu 4
Oeu 5
Oeu 6
Oeu 7
Oeu 8
Oeu 9
Oeu 10
Oeu 11
Oeu 12
Oeu 13
Oeu 14
Oeu 15
Oeu 16
Oeu 17
Oeu 18
Oeu 19
Oeu 20
Oeu 21
Oeu 22
Oeu 23