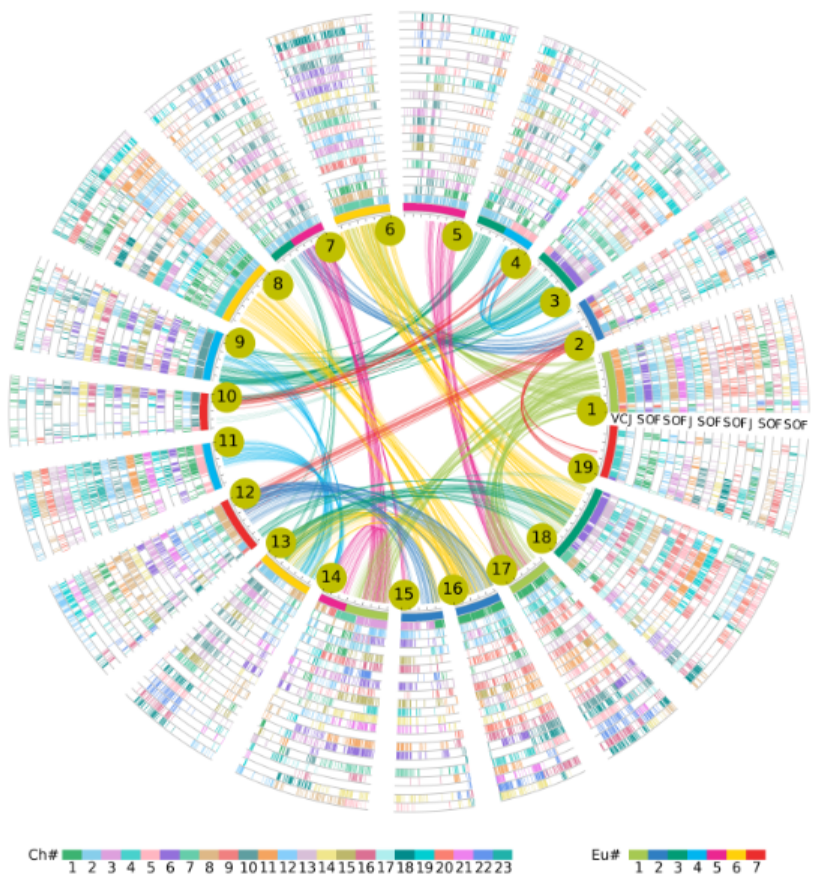
Hierarchical genome alignment list
To present identified gene homology information associated with evolutionary events, we constructed Hierarchical genome alignment list (HGAL) to store multigenome collinear genes across Oleaceae using Vitis vinifera as the reference genome. In the HGAL, all V. vinifera genes are placed in the first column. Due to the ECH event that triplicated the ancestral eudicot genome, each gene in V. vinifera has two paralogs. Thus, we used two additional columns to accommodate the paralogs. For each V. vinifera gene, if an orthologous gene appears in the expected genomic location of Oleaceae species, the gene ID is entered into the corresponding cell. When an orthologous gene is missing, often due to the absence or translocation of a gene in the genome, a dot is placed in the cell. If a specific Oleaceae genome has undergone additional rounds of tetraploidization events after the ACH, we can assign 3xNx2 columns for this Oleaceae plant.